SOM Visualization Guide
This comprehensive guide covers all visualization capabilities available in TorchSOM for analyzing and interpreting Self-Organizing Maps effectively.
Overview
TorchSOM provides a rich set of visualization tools through the SOMVisualizer class, supporting both rectangular and hexagonal topologies. All visualizations are designed to help you understand:
Training Progress: How well your SOM is learning over time
Data Distribution: How input data maps onto the SOM grid
Topology Preservation: Whether neighborhood relationships are maintained
Feature Representation: How individual features are distributed across neurons
Cluster Structure: Identification of natural groupings in your data
Quick Start
Basic Visualization Setup
from torchsom import SOM
from torchsom.visualization import SOMVisualizer, VisualizationConfig
import torch
# Train a SOM
data = torch.randn(1000, 4)
som = SOM(x=20, y=15, num_features=4, epochs=50)
som.initialize_weights(data=data, mode="pca")
q_errors, t_errors = som.fit(data)
# Create visualizer with default configuration
visualizer = SOMVisualizer(som=som)
# Generate all visualizations at once
visualizer.plot_all(
quantization_errors=q_errors,
topographic_errors=t_errors,
data=data,
save_path="som_results"
)
Custom Configuration
The VisualizationConfig class provides comprehensive customization options:
config = VisualizationConfig(
figsize=(12, 8), # Figure size in inches
fontsize={ # Font sizes for different elements
"title": 16,
"axis": 13,
"legend": 11
},
fontweight={ # Font weights
"title": "bold",
"axis": "normal"
},
cmap="viridis", # Default colormap
dpi=150, # Resolution for saved figures
grid_alpha=0.3, # Grid transparency
colorbar_pad=0.01, # Colorbar padding
save_format="png", # Save format (png, pdf, eps, svg)
hexgrid_size=None # Hexagonal grid size (auto if None)
)
Visualization Types
Training Errors
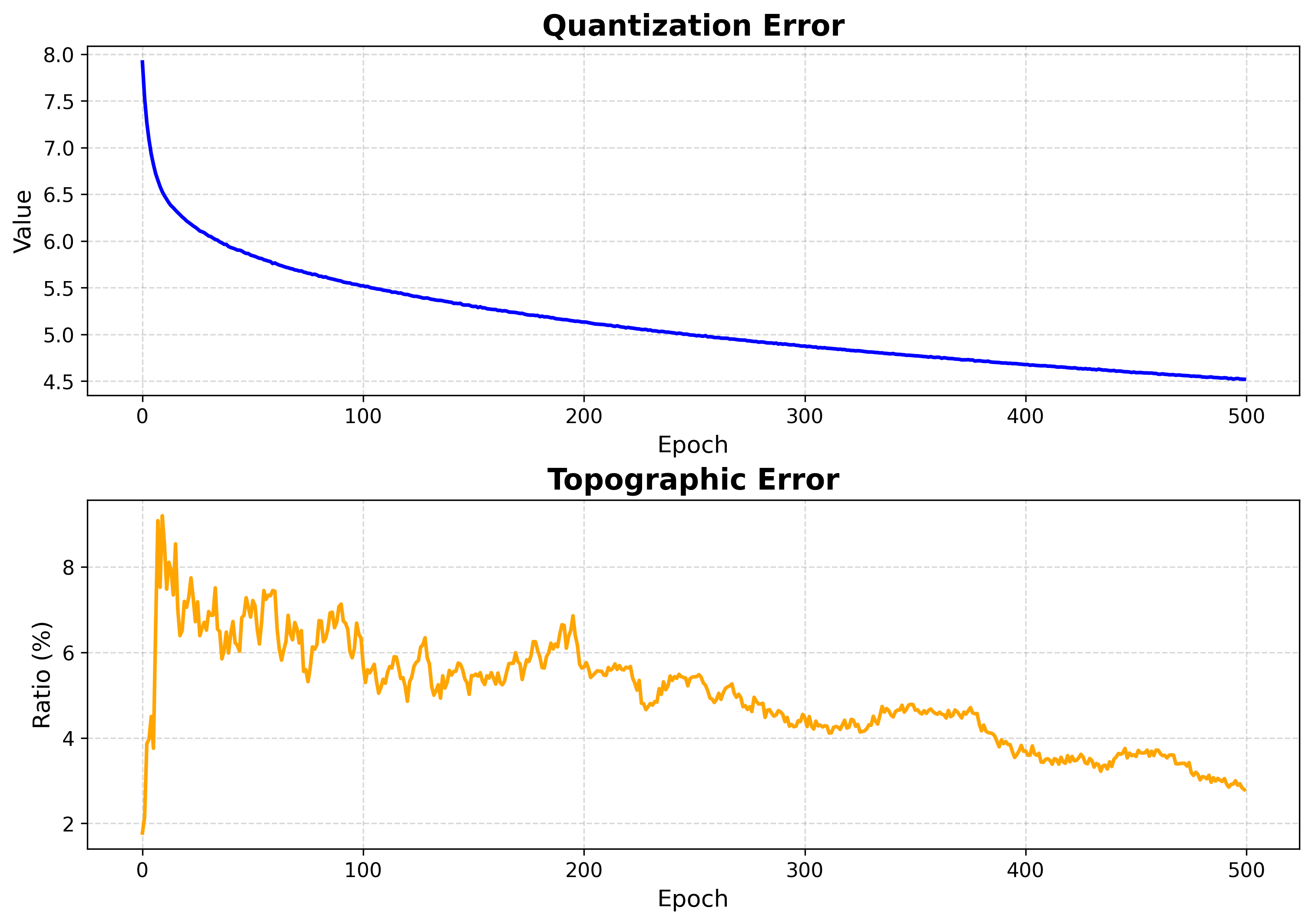
Monitors SOM learning progress by plotting quantization and topographic errors over epochs.
visualizer.plot_training_errors(
quantization_errors=q_errors,
topographic_errors=t_errors,
save_path="results"
)
Interpretation:
Quantization Error: Measures how well the SOM represents the input data (lower is better)
Topographic Error: Measures topology preservation (lower percentage is better)
Convergence: Both errors should generally decrease and stabilize during training
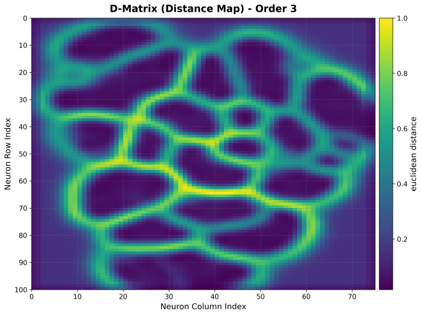
Distance Map (U-Matrix)
The unified distance matrix shows the distance between each neuron and its neighbors, revealing cluster boundaries.
visualizer.plot_distance_map(
save_path=save_path
distance_metric=som.distance_fn_name,
neighborhood_order=som.neighborhood_order,
scaling="sum",
)
Interpretation:
Dark Regions: Small distances between neighboring neurons (cluster boundaries)
Light Regions: Large distances between neighboring neurons (within clusters)
Topology: Works with both rectangular and hexagonal grids
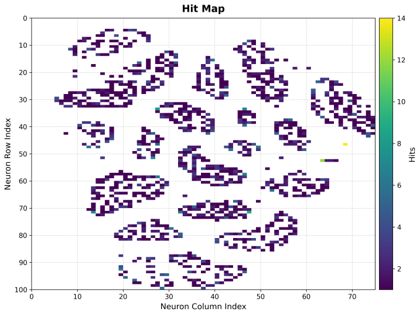
Hit Map
Shows the frequency of neuron activation, indicating how often each neuron was selected as the Best Matching Unit (BMU).
visualizer.plot_hit_map(
data=train_features,
save_path=save_path,
batch_size=train_features.shape[0],
)
Interpretation:
Bright Areas: Frequently activated neurons (high data density)
Dark Areas: Rarely activated neurons (low data density or dead neurons)
Usage: Identifies data distribution patterns and potential dead neurons
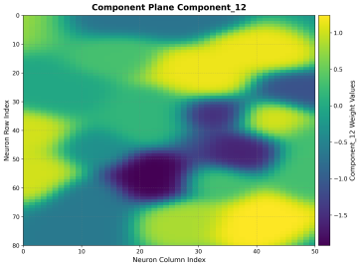
Component Planes
Individual visualizations for each input feature dimension, showing how feature weights are distributed across the map.
visualizer.plot_component_planes(
component_names=feature_names,
save_path=save_path
)
Interpretation:
One Plane per Feature: Shows weight values for each input dimension
Pattern Analysis: Reveals feature level in different map regions
Supervised Maps
Visualizations for supervised learning tasks, including both classification and regression, help interpret how target information is distributed across the SOM map.
Classification Case
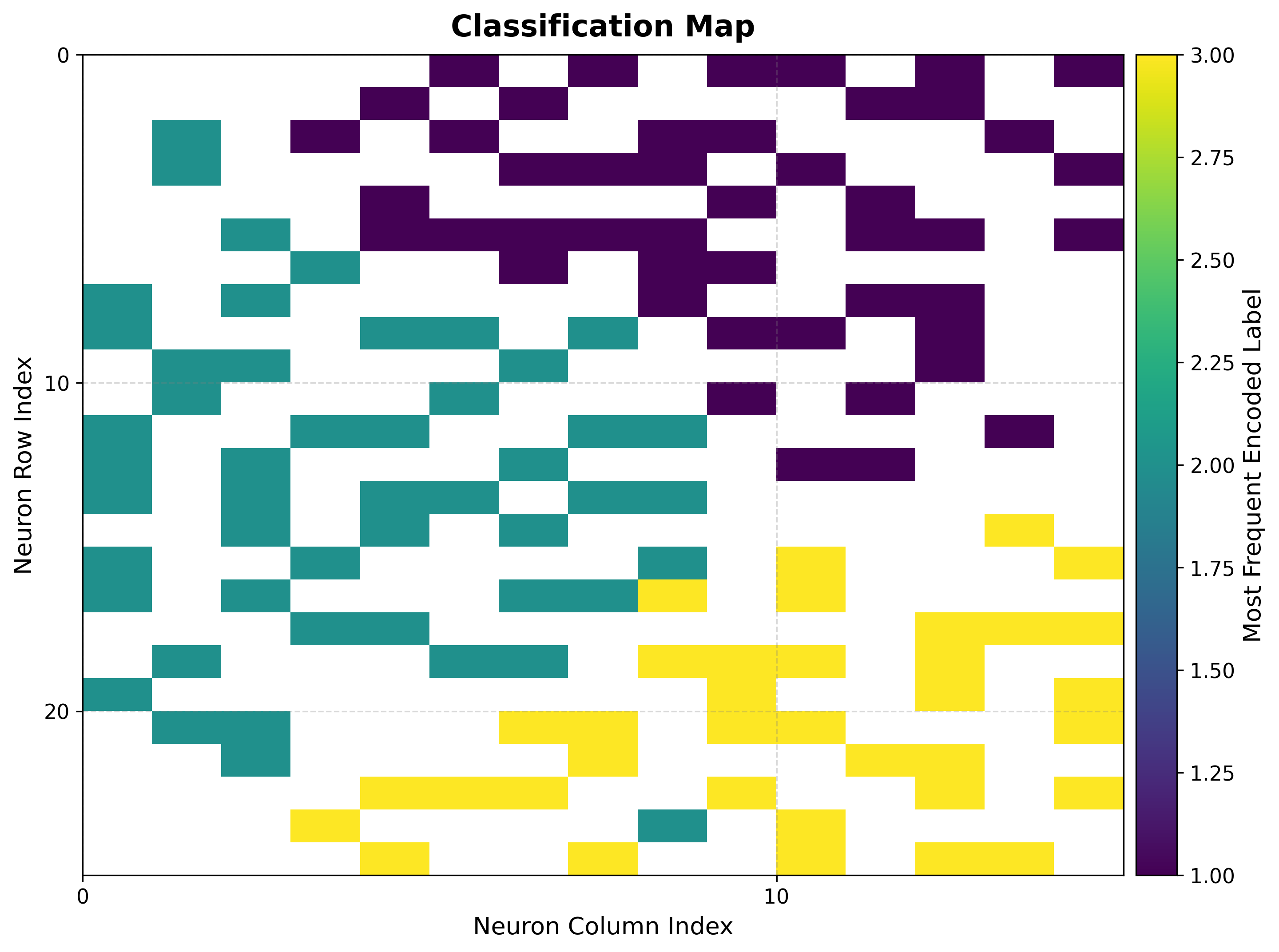
Displays the most frequent class label assigned to each neuron, providing insight into class separation and cluster structure.
visualizer.plot_classification_map(
data=train_features,
target=train_targets,
save_path=save_path,
bmus_data_map=bmus_map,
neighborhood_order=som.neighborhood_order,
)
Interpretation:
Color Coding: Each color represents a different class label.
Cluster Identification: Reveals spatial organization of classes on the map.
Decision Boundaries: Boundaries between colors indicate class separation.
Regression Case
Analyzes the distribution of continuous target values (e.g., for regression tasks) using statistical summaries per neuron.
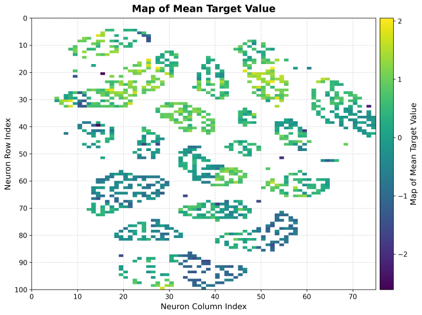
Mean Map
Shows the average target value for samples mapped to each neuron.
visualizer.plot_metric_map(
data=train_features,
target=train_targets,
reduction_parameter="mean",
save_path=save_path,
bmus_data_map=bmus_map,
)
Interpretation:
Color Scale: Indicates the mean target value per neuron.
Smooth Transitions: Suggest good topology preservation.
Hot Spots: Highlight neurons with extreme target values.
Standard Deviation Map
Shows the variability of target values for each neuron, useful for assessing prediction reliability.
visualizer.plot_metric_map(
data=train_features,
target=train_targets,
reduction_parameter="std",
save_path=save_path,
bmus_data_map=bmus_map,
)
Interpretation:
Low Values: Neurons with consistent (low-variance) target values—good for prediction.
High Values: Neurons with variable (high-variance) target values—less reliable.
Quality Assessment: Helps identify the most reliable neurons for regression tasks.
Advanced Visualizations
Score Map
Evaluates neuron representativeness using a composite score combining standard error and sample distribution.
visualizer.plot_score_map(
bmus_data_map=bmus_map,
target=train_targets,
total_samples=train_features.shape[0],
save_path=save_path,
)
where:
\(S_{ij} \in \mathbb{R}^+\): Reliability score of neuron at position \((i, j)\)
\(\sigma_{ij} \in \mathbb{R}^+\): Standard deviation of target values assigned to neuron at position \((i, j)\)
\(n_{ij} \in \mathbb{N}\): Number of samples assigned to neuron at position \((i, j)\)
\(N \in \mathbb{N}\): Total number of samples in the latent space
Interpretation:
Lower Scores: Better neuron representativeness
Usage: Identifies most reliable neurons for analysis
Rank Map
Ranks neurons based on their target value standard deviations.
visualizer.plot_rank_map(
bmus_data_map=bmus_map,
target=train_targets,
save_path=save_path,
)
Interpretation:
Rank 1: Lowest standard deviation (best predictive neurons)
Higher Ranks: Increasing standard deviation
Selection: Use top-ranked neurons for reliable predictions
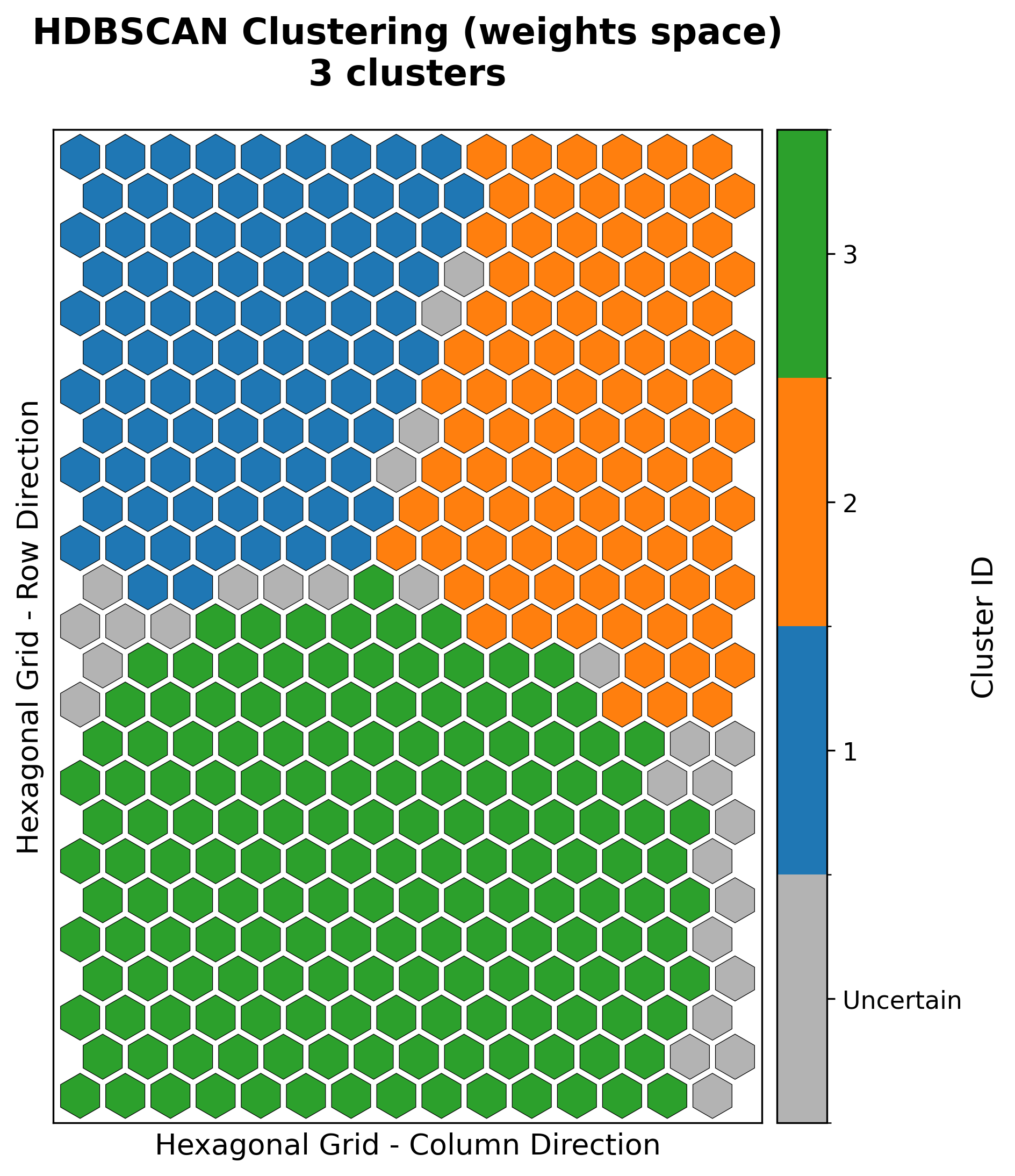
Cluster Map
Clusters neurons based on algorithms like HDBSCAN, KMeans, or GMMs.
cluster = som.cluster(
method="hdbscan", # hdbscan, kmeans, gmm
n_clusters=n_clusters,
feature_space="weights",
)
visualizer.plot_cluster_map(
cluster_result=cluster,
save_path=save_path,
)
Troubleshooting
- White Cells in Visualizations:
Indicates neurons with zero values or NaN
Check for dead neurons in hit map
Verify data preprocessing and normalization
- Memory Issues:
Reduce batch size in visualization functions
Use CPU-only mode for very large SOMs
Clear GPU cache with
torch.cuda.empty_cache()
- Topology Preservation:
High topographic error indicates poor topology preservation
Consider adjusting learning rate, sigma, or training epochs
Use PCA initialization for better convergence
References
For more examples and detailed usage, see: